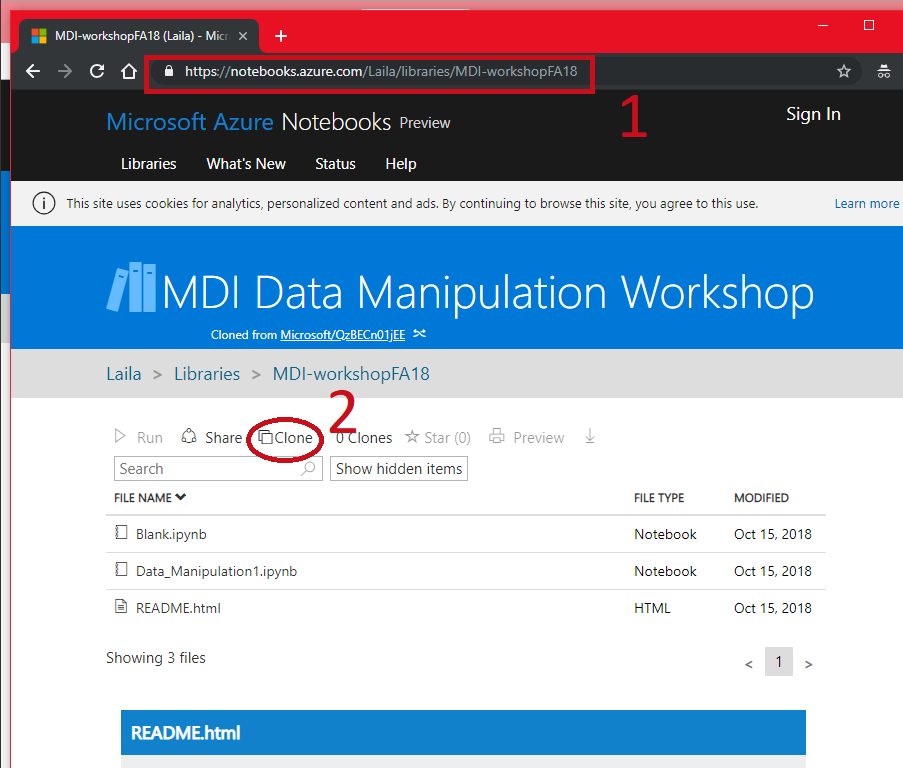
Follow Along¶
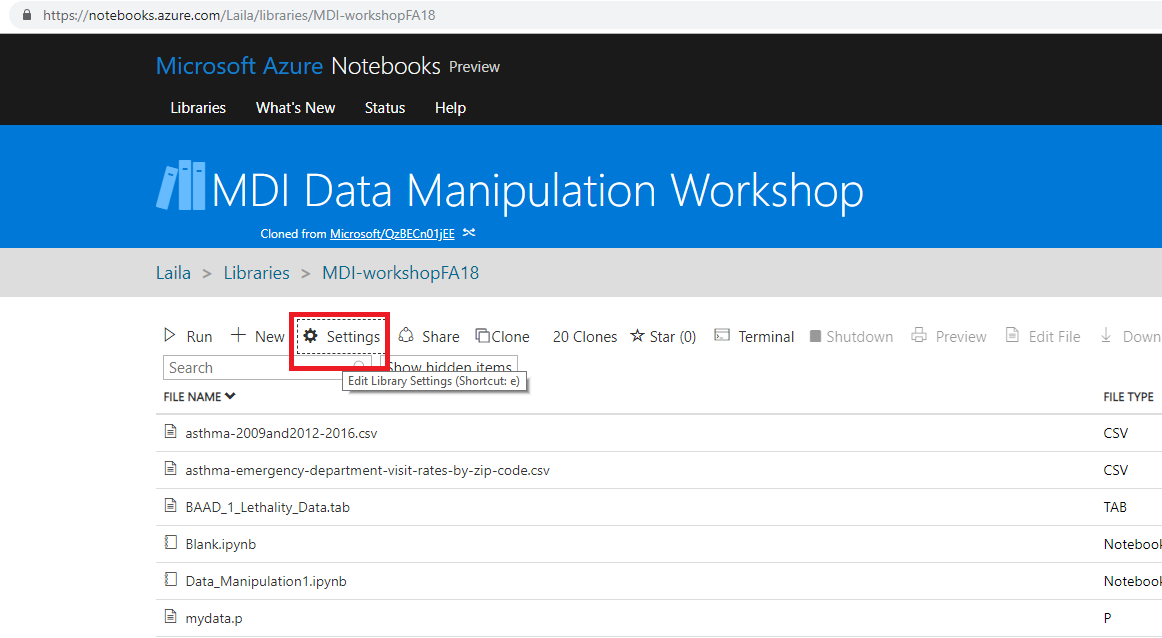
- Go to https://notebooks.azure.com/Laila/libraries/MDI-workshopFA18
- Clone the directory
Follow Along¶
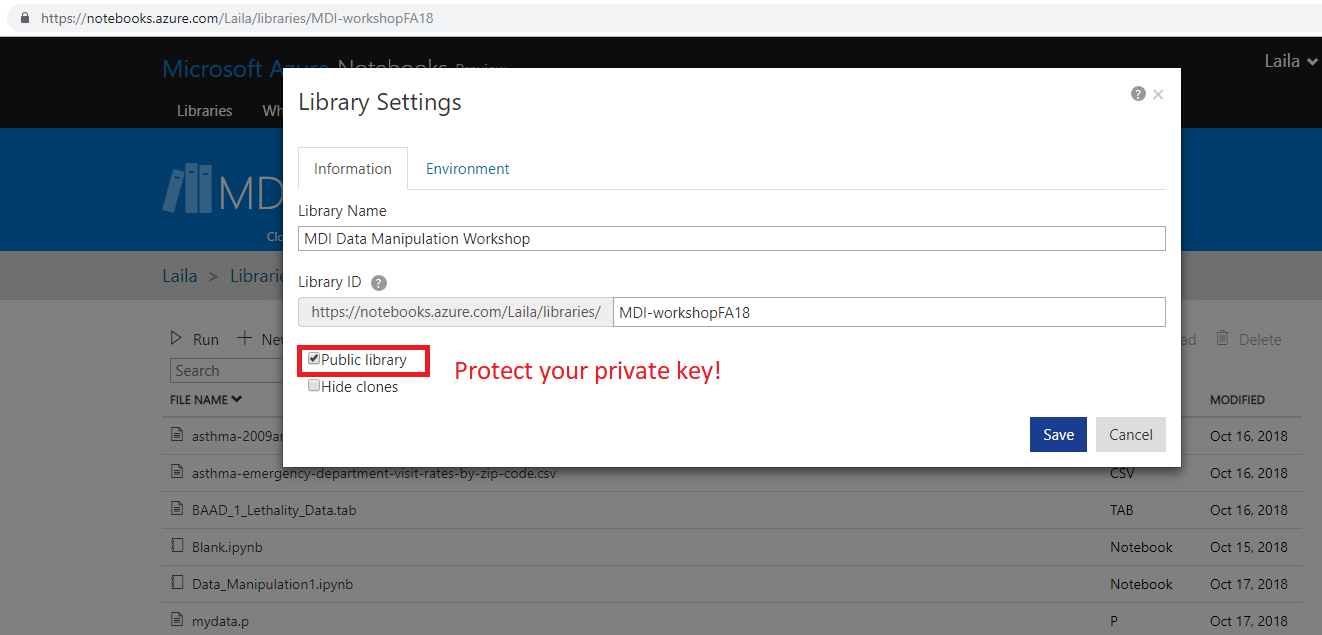
- Sign in with any Microsoft Account (Hotmail, Outlook, Azure, etc.)
- Create a folder to put it in, mark as private or public
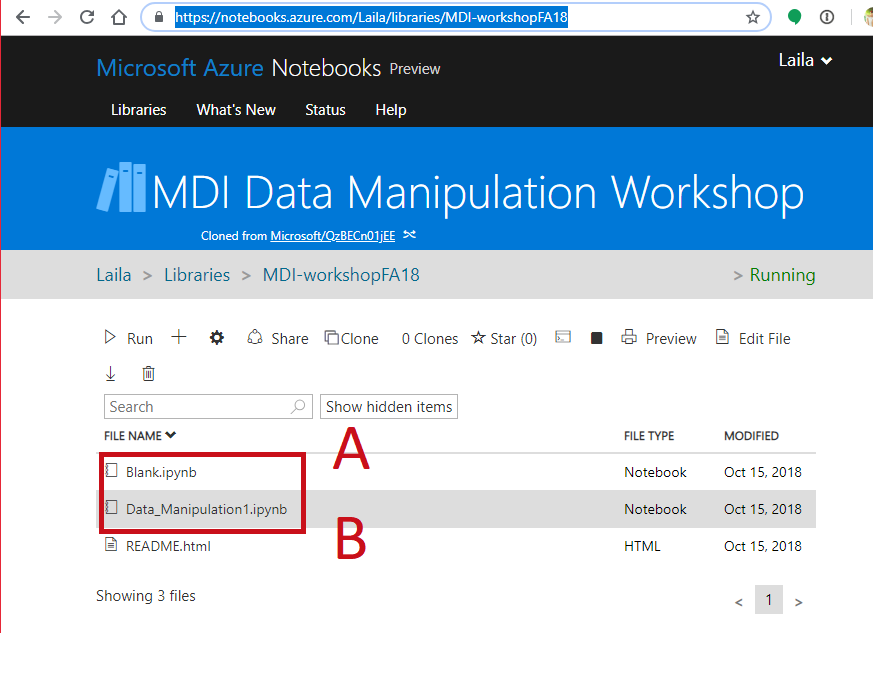
Follow Along¶
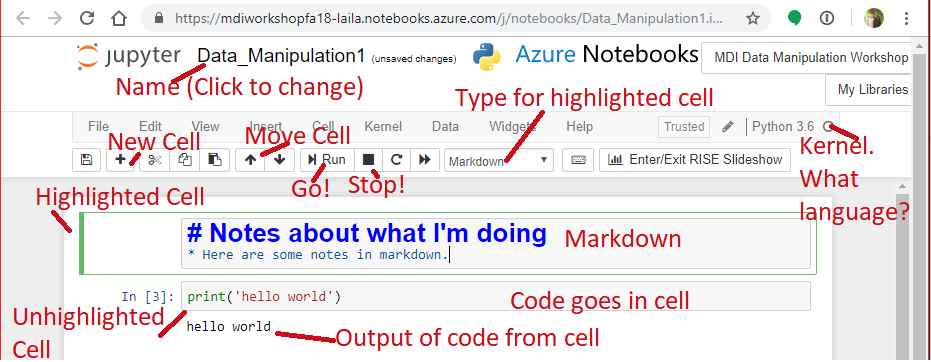
- Open a notebook
- Open this notebook to have the code to play with
- Data_collection_Analysis2.ipynb
- Open a blank notebook to follow along and try on your own.
Your Environment¶
- Jupyter Notebook Hosted in Azure
- Want to install it at home?
- Install the Anaconda distribution of Python https://www.anaconda.com/download/
- Install Jupyter Notebooks http://jupyter.org/install
Agenda for today:¶
- Collect data from APIs
- Scrape data
- Merge data into a data frame
- Statsmodels package
- SKLearn package
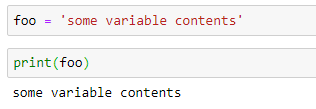
Packages to Import For Today¶
- Should all be included with your Anaconda Python Distribution
- Raise your hand for help if you have trouble
- Our plots will use matplotlib, similar to plotting in matlab
- %matplotlib inline tells Jupyter Notebooks to display your plots
- from allows you to import part of a package
In [1]:
import pandas as pd
import numpy as np
import pickle
import statsmodels.api as sm
from sklearn import cluster
import matplotlib.pyplot as plt
%matplotlib inline
from bs4 import BeautifulSoup as bs
import requests
import time
# from ggplot import *
Other Useful Packages (not used today)¶
- ggplot: the familiar ggplot2 you know and love from R
- seaborn: Makes your plots prettier
- plotly: makes interactive visualizations, similar to shiny
- gensim: package for doing natural language processing
- scipy: used with numpy to do math. Generates random numbers from distributions, does matrix operations, etc.
Scraping¶
How the Internet Works¶
- Code is stored on servers
- Web addresses point to the location of that code
- Going to an address or clicking a button sends requests to the server for data,
- The server returns the requested content
- Your web browser interprets the code to render the web page
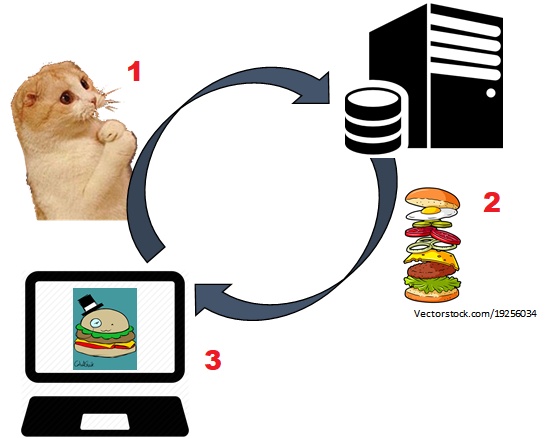
Scraping:¶
- Collect the website code by emulating the process:
- Can haz cheezburger?

- Can haz cheezburger?
- Extract the useful information from the scraped code:
- Where's the beef?

- Where's the beef?
API¶
Application Programming Interface¶
- The set of rules that govern communication between two pieces of code
- Code requires clear expected inputs and outputs
- APIs define required inputs to get the outputs in a format you can expect.
- Easier than scraping a website because gives you exactly what you ask for

API Keys¶
APIs often require identification¶
- Go to https://docs.airnowapi.org
- Register and get a key
- Log in to the site
- Select web services
DO NOT SHARE YOUR KEY¶
- It will get stolen and used for malicious activity
Requests to a Server¶
GET
- Requests data from the server
- Encoded into the URL

POST
- Submits data to be processed by the server
- For example, filter the data
- Can attach additional data not directly in the url

Requests encoded in the URL¶
Parsing a URL¶
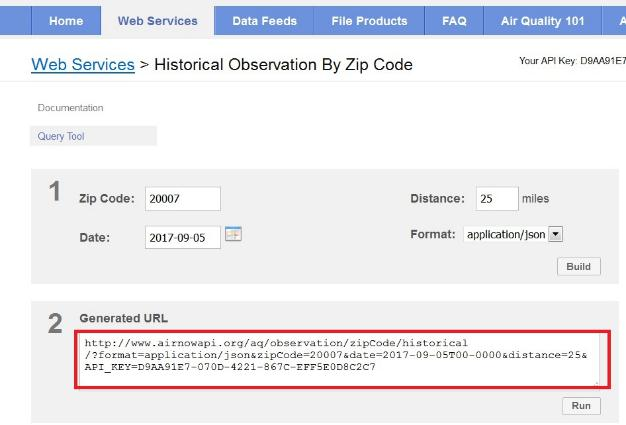
http://www.airnowapi.org/aq/observation/zipCode/historical/?
format=application/json&
zipCode=20007&
date=2017-09-05T00-0000&
distance=25&
API_KEY=D9AA91E7-070D-4221-867CC-XXXXXXXXXXX
The base URL or endpoint is:
http://www.airnowapi.org/aq/observation/zipCode/historical/? tells us that this is a query.
& separates name, value pairs within the request.
Five name, value pairs POSTED
- format, zipCode, date, distance, API_KEY
In [3]:
base_url = "http://www.mywebsite.com/data/api?"
attributes = ["key1=value1",
"key2=value2",
"API_KEY=39DC3727-09BD-XXXX-XXXX-XXXXXXXXXXXX"
]
post_url = '&'.join(attributes)
print(base_url+post_url)
http://www.mywebsite.com/data/apikey1=value1&key2=value2&API_KEY=39DC3727-09BD-XXXX-XXXX-XXXXXXXXXXXX
Prepare another URL on your own¶
In [4]:
base_url = "http://www.airnowapi.org/aq/observation/zipCode/historical/"
attributes = ["format=application/json",
"zipCode=20007",
"date=2017-09-05T00-0000",
"distance=25",
"API_KEY=39DC3727-09BD-48C4-BBD8-XXXXXXXXXXXX"
]
post_url = '&'.join(attributes)
print(base_url+post_url)
http://www.airnowapi.org/aq/observation/zipCode/historical/format=application/json&zipCode=20007&date=2017-09-05T00-0000&distance=25&API_KEY=39DC3727-09BD-48C4-BBD8-XXXXXXXXXXXX
Where did the ? go?¶
Requests from Python¶
- Use requests package
- .get(url,post) for get and post requests
- First parameter: url for get request
- Optional second parameter: post data if doing a post request
- Requests posts the data to the url, so you don't need to add a ? to the base url
- Leave the post data out for a get request instead of a post request
- Requested json format
- Returns list of dictionaries
- Look at the returned keys
In [17]:
ingredients=requests.get(base_url, post_url)
ingredients = ingredients.json()
print(ingredients[0])
{'DateObserved': '2017-09-05 ', 'HourObserved': 0, 'LocalTimeZone': 'EST', 'ReportingArea': 'Metropolitan Washington', 'StateCode': 'DC', 'Latitude': 38.919, 'Longitude': -77.013, 'ParameterName': 'OZONE', 'AQI': 47, 'Category': {'Number': 1, 'Name': 'Good'}}
View Returned Data:¶
- Each list gives a different parameter for zip code and date we searched
In [18]:
for item in ingredients:
AQIType = item['ParameterName']
City=item['ReportingArea']
AQIValue=item['AQI']
print("For Location ", City, " the AQI for ", AQIType, "is ", AQIValue)
For Location Metropolitan Washington the AQI for OZONE is 47 For Location Metropolitan Washington the AQI for PM2.5 is 61 For Location Metropolitan Washington the AQI for PM10 is 13
Ethics¶
- Check the websites terms of use
- Don't hit too hard:
- Insert pauses in your code to act more like a human
- Scraping can look like an attack
- Server will block you without pauses
- APIs often have rate limits
- Use the time package to pause for a second between hits
In [ ]:
time.sleep(1)
Review from last week: Asthma Data¶
- Load asthma data from csv
- Display a few rows
- Fix the zip codes
- Pivot to have one row per zip code (repeated rows for children, adults, all)
Load from csv, display¶
In [7]:
asthma_data = pd.read_csv('asthma-emergency-department-visit-rates-by-zip-code.csv')
asthma_data.head(2)
Out[7]:
| Year | ZIP code | Age Group | Number of Visits | Age-adjusted rate | County Fips code | County | |
|---|---|---|---|---|---|---|---|
| 0 | 2015 | 90004\n(34.07646, -118.309453) | Children (0-17) | 117.0 | 91.7 | 6037 | LOS ANGELES |
| 1 | 2015 | 90011\n(34.007055, -118.258872) | Children (0-17) | 381.0 | 102.8 | 6037 | LOS ANGELES |
Fix the zip codes¶
- Fix zip codes
In [8]:
asthma_data[['zip','coordinates']] = asthma_data.loc[:,'ZIP code'].str.split(
pat='\n',expand=True)
asthma_data.drop('ZIP code', axis=1,inplace=True)
asthma_data.head(2)
Out[8]:
| Year | Age Group | Number of Visits | Age-adjusted rate | County Fips code | County | zip | coordinates | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2015 | Children (0-17) | 117.0 | 91.7 | 6037 | LOS ANGELES | 90004 | (34.07646, -118.309453) |
| 1 | 2015 | Children (0-17) | 381.0 | 102.8 | 6037 | LOS ANGELES | 90011 | (34.007055, -118.258872) |
Pivot the data so age group are columns¶
In [9]:
asthma_unstacked = asthma_data.pivot_table(index = ['Year',
'zip',
'County',
'coordinates',
'County Fips code'],
columns = 'Age Group',
values = 'Number of Visits')
asthma_unstacked.reset_index(drop=False,inplace=True)
asthma_unstacked.head(2)
Out[9]:
| Age Group | Year | zip | County | coordinates | County Fips code | Adults (18+) | All Ages | Children (0-17) |
|---|---|---|---|---|---|---|---|---|
| 0 | 2009 | 90001 | LOS ANGELES | (33.973252, -118.249154) | 6037 | 206.0 | 409.0 | 203.0 |
| 1 | 2009 | 90002 | LOS ANGELES | (33.949079, -118.247877) | 6037 | 204.0 | 418.0 | 214.0 |
Now we have some zip codes!¶
Automate Data Collection¶
- Request the data for those zipcodes on a day in 2015 (you pick, fire season July-Oct)
- Be sure to sleep between requests
- Store that data as you go into a dictionary
- Key: zip code
- Value: Dictionary of the air quality parameters and their value
In [ ]:
base_url = "http://www.airnowapi.org/aq/observation/zipCode/historical/"
zips = asthma_unstacked.zip.unique()
zips = zips[:450]
date ="date=2015-09-01T00-0000"
api_key = "API_KEY=39DC3727-09BD-48C4-BBD8-XXXXXXXXXXXX"
return_format = "format=application/json"
zip_str = "zipCode="
post_url = "&".join([date,api_key,return_format,zip_str])
data_dict = {}
for zipcode in zips:
time.sleep(1)
zip_post = post_url + str(zipcode)
ingredients = requests.get(base_url, zip_post)
ingredients = ingredients.json()
zip_data = {}
for data_point in ingredients:
AQIType = data_point['ParameterName']
AQIVal = data_point['AQI']
zip_data[AQIType] = AQIVal
data_dict[zipcode]= zip_data
Scraping: Parsing HTML¶
- What about when you don't have an API that returns dictionaries?
- HTML is a markup language that displays data (text, images, etc)
- Puts content within nested tags to tell your browser how to display it
<Section_tag>¶
<tag> Content </tag>
<tag> Content </tag>
< /Section_tag>¶
<Section_tag>¶
<tag> Beef </tag>
< /Section_tag>¶
Find the tags that identify the content you want:¶
- First paragraph of wikipedia article: https://en.wikipedia.org/wiki/Data_science
- Inspect the webpage:
- Windows: ctrl+shift+i
- Mac: ctrl+alt+i
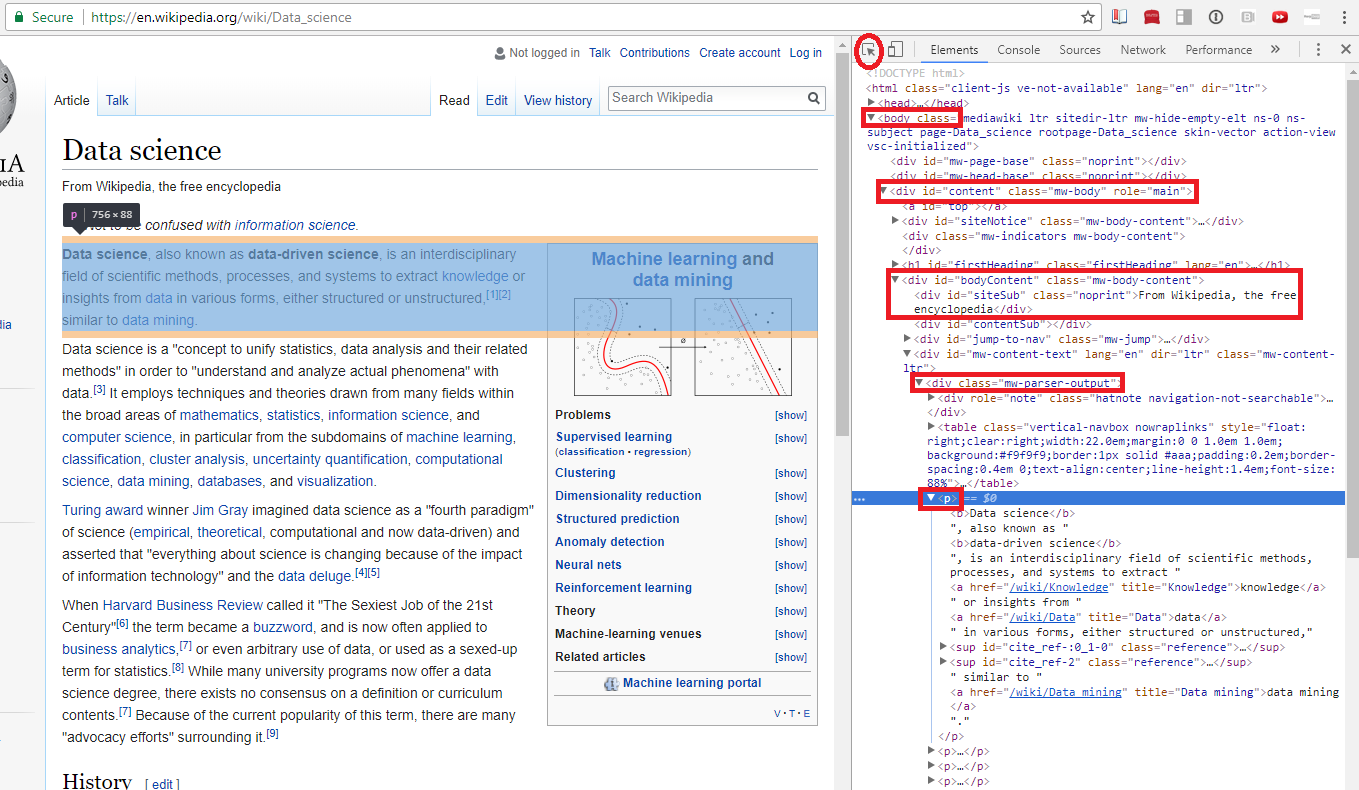
Parsing HTML with Beautiful Soup¶
Beautiful Soup takes the raw html and parses the tags so you can search through them.¶
- text attribute returns raw html text from requests
- Ignore the warning, default parser is fine
- We know it's the first paragraph tag in the body tag, so:
- Can find first tag of a type using .
- What went wrong?
In [24]:
ingredients = requests.get("https://en.wikipedia.org/wiki/Data_science")
soup = bs(ingredients.text)
print(soup.body.p)
<p class="mw-empty-elt"> </p>
Use Find Feature to Narrow Your Search¶
- Find the unique div we identified
- Remember the underscore: "class_"
- Find the p tag within the resulting html
- Use an index to return just the first paragraph tag
- Use the text attribute to ignore all the formatting and link tags
In [15]:
parser_div = soup.find("div", class_="mw-parser-output")
wiki_content = parser_div.find_all('p')
print(wiki_content[1])
print('*****************************************')
print(wiki_content[1].text)
<p><b>Data science</b> is an <a class="mw-redirect" href="/wiki/Interdisciplinary" title="Interdisciplinary">interdisciplinary</a> field that uses scientific methods, processes, algorithms and systems to extract <a href="/wiki/Knowledge" title="Knowledge">knowledge</a> and insights from <a href="/wiki/Data" title="Data">data</a> in various forms, both structured and unstructured,<sup class="reference" id="cite_ref-:0_1-0"><a href="#cite_note-:0-1">[1]</a></sup><sup class="reference" id="cite_ref-2"><a href="#cite_note-2">[2]</a></sup> similar to <a href="/wiki/Data_mining" title="Data mining">data mining</a>. </p> ***************************************** Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.
List Contents¶
In [28]:
parser_div = soup.find("div", id="toc")
wiki_content = parser_div.find_all('ul')
for item in wiki_content:
print(item.text)
1 History 2 Relationship to statistics 3 See also 4 References
Get All Links in the History Section¶
- Hint: chrome's inspect replaces "&" with "&" in links
In [56]:
wiki_content = soup.find_all('a',href=True)
in_hist = False
links = []
for l in wiki_content:
link = l['href']
if link == '/w/index.php?title=Data_science&action=edit§ion=2':
in_hist = False
if in_hist:
links.append(link)
if link =="/w/index.php?title=Data_science&action=edit§ion=1":
in_hist = True
print(links)
['/wiki/Computer_science', '/wiki/Peter_Naur', '/wiki/Datalogy', '#cite_note-15', '#cite_note-16', '#cite_note-Hayashi-3', '/wiki/C.F._Jeff_Wu', '#cite_note-cfjwutk-17', '/wiki/University_of_Michigan', '#cite_note-cfjwu01-18', '#cite_note-cfjwutk-17', '#cite_note-cfjwu02-19', '/wiki/Prasanta_Chandra_Mahalanobis', '/wiki/Indian_Statistical_Institute', '#cite_note-cleveland01-20', '#cite_note-ics12-21', '#cite_note-dsj12-22', '#cite_note-dsj02-23', '#cite_note-jds03-24', '#cite_note-25', '/wiki/Wikipedia:Citation_needed', '/wiki/Jim_Gray_(computer_scientist)', '#cite_note-TansleyTolle2009-4', '#cite_note-BellHey2009-5', '#cite_note-26', '/wiki/Harvard_Business_Review', '#cite_note-Harvard-6', '/wiki/DJ_Patil', '/wiki/Jeff_Hammerbacher', '#cite_note-27', 'http://euads.org', '#cite_note-28', '/wiki/General_Assembly_(school)', '/wiki/The_Data_Incubator', '#cite_note-29', '/wiki/American_Statistical_Association', '#cite_note-ASA-30', '#cite_note-31', 'http://www.gfkl.org/welcome/', '/wiki/University_of_Essex']
Use a for loop and scrape the first paragraph from a bunch of wikipedia articles¶
- Add your own subjects
In [20]:
topics = ['Data_scraping','Machine_learning','Statistics','Linear_algebra',
'Cluster_analysis','Scientific_modelling','Analysis','Linear_regression']
base_url = 'https://en.wikipedia.org/wiki/'
paragraphs = []
for topic in topics:
url = base_url.format(topic)
ingredients = requests.get(base_url+topic)
soup = bs(ingredients.text)
parser_div = soup.find("div", class_="mw-parser-output")
wiki_content = parser_div.find_all('p')
for p in range(10):
if len(wiki_content[p].text)>10:
paragraphs.append(wiki_content[p].text)
break
time.sleep(1)
print(dict(zip(topics,paragraphs)))
{'Data_scraping': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Machine_learning': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Statistics': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Linear_algebra': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Cluster_analysis': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Scientific_modelling': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Analysis': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n', 'Regression': 'Data science is an interdisciplinary field that uses scientific methods, processes, algorithms and systems to extract knowledge and insights from data in various forms, both structured and unstructured,[1][2] similar to data mining.\n'}
Back To Our Data¶
- If it's still running, go ahead and stop it by pushing the square at the top of the notebook:
- Save what you collected, don't want to hit them twice!
In [61]:
pickle.dump(data_dict,open('AQI_data_raw.p','wb'))
Subset down to the data we have:¶
- use the isin() method to include only those zip codes we've already collected
In [63]:
collected = list(data_dict.keys())
asthma_2015_sub = asthma_unstacked.loc[(asthma_unstacked.zip.isin(collected))&
(asthma_unstacked.Year == 2015),:]
Create a dataframe from the new AQI data¶
In [184]:
aqi_data = pd.DataFrame.from_dict(data_dict, orient='index')
aqi_data.reset_index(drop=False,inplace=True)
aqi_data.rename(columns={'index':'zip'},inplace=True)
aqi_data.head()
Out[184]:
| zip | OZONE | day | PM2.5 | PM10 | |
|---|---|---|---|---|---|
| 0 | 90001 | 36.0 | 1 | NaN | NaN |
| 1 | 90002 | 36.0 | 1 | NaN | NaN |
| 2 | 90003 | 36.0 | 1 | NaN | NaN |
| 3 | 90004 | 54.0 | 1 | NaN | NaN |
| 4 | 90005 | 54.0 | 1 | NaN | NaN |
Combine The Data¶
https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.merge.html
- Types of merges:
- Left: Use only rows from the dataframe you are merging into
- Right: use only rows from the dataframe you are inserting, (the one in the parentheses)
- Inner: Use only rows that match between both
- Outer: Use all rows, even if they only appear in one of the dataframes
- On: The variables you want to compare
- Specify right_on and left_on if they have different names
In [207]:
asthma_aqi = asthma_2015_sub.merge(aqi_data,how='outer',on='zip')
asthma_aqi.rename(columns = {'Adults (18+)':'Adults',
'All Ages':'Incidents',
'Children (0-17)':'Children'},inplace=True)
asthma_aqi.head(2)
Out[207]:
| Year | zip | County | coordinates | County Fips code | Adults | Incidents | Children | OZONE | day | PM2.5 | PM10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2015.0 | 90001 | LOS ANGELES | (33.973252, -118.249154) | 6037.0 | 229.0 | 441.0 | 212.0 | 36.0 | 1.0 | NaN | NaN |
| 1 | 2015.0 | 90002 | LOS ANGELES | (33.949079, -118.247877) | 6037.0 | 249.0 | 476.0 | 227.0 | 36.0 | 1.0 | NaN | NaN |
Look At The Data: Histogram¶
- 20 bins
In [186]:
asthma_aqi.Incidents.plot.hist(20)
Out[186]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f05db5d3198>
Look At The Data: Smoothed Distribution¶
In [187]:
asthma_aqi.loc[:,['Incidents','OZONE']].plot.density()
Out[187]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f05dc788748>
Look at particulates¶
- There is a lot of missingness in 2015
- Try other variables, such as comparing children and adults
In [188]:
asthma_aqi.loc[:,['PM2.5','PM10']].plot.hist()
Out[188]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f05d40b7c18>
Scatter Plot¶
- Try some other combinations
- Our data look clustered, but we'll ignore that for now
In [189]:
asthma_aqi.plot.scatter('OZONE','PM2.5')
Out[189]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f05d40bfa58>
Run a regression:¶
- Note: statsmodels supports equation format like R
http://www.statsmodels.org/dev/example_formulas.html
In [200]:
y =asthma_aqi.loc[:,'Incidents']
x =asthma_aqi.loc[:,['OZONE','PM2.5']]
x['c'] = 1
ols_model1 = sm.OLS(y,x,missing='drop')
results = ols_model1.fit()
print(results.summary())
pickle.dump([results,ols_model1],open('ols_model_results.p','wb'))
OLS Regression Results
==============================================================================
Dep. Variable: Incidents R-squared: 0.025
Model: OLS Adj. R-squared: 0.001
Method: Least Squares F-statistic: 1.022
Date: Thu, 25 Oct 2018 Prob (F-statistic): 0.364
Time: 13:36:39 Log-Likelihood: -523.77
No. Observations: 83 AIC: 1054.
Df Residuals: 80 BIC: 1061.
Df Model: 2
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
OZONE -6.8458 6.033 -1.135 0.260 -18.851 5.159
PM2.5 -1.3005 2.426 -0.536 0.593 -6.128 3.527
c 485.5302 236.518 2.053 0.043 14.844 956.216
==============================================================================
Omnibus: 24.776 Durbin-Watson: 1.271
Prob(Omnibus): 0.000 Jarque-Bera (JB): 34.046
Skew: 1.400 Prob(JB): 4.05e-08
Kurtosis: 4.415 Cond. No. 1.01e+03
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 1.01e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
Evaluate the model with some regression plots¶
Partial Regressions to see effect of each variable¶
In [191]:
fig = plt.figure(figsize=(12,8))
fig = sm.graphics.plot_partregress_grid(results, fig=fig)
Population confound¶
- Fires spread in less populated areas
- Fewer people to have asthma attacks in less populated areas
- Collect population data
- Use pandas to read the html table directly
In [208]:
ingredients = requests.get('https://www.california-demographics.com/zip_codes_by_population')
soup = bs(ingredients.text)
table = soup.find("table")
population = pd.read_html(str(table),flavor='html5lib')[0]
population.rename(columns=population.iloc[0],inplace=True)
population.drop(labels=0, axis=0,inplace=True)
population.head(2)
Out[208]:
| California Zip Codes by Population Rank | Zip Code | Population | |
|---|---|---|---|
| 1 | 1 | 90650 | 106360 |
| 2 | 2 | 90011 | 104762 |
Fix zipcode column¶
- Split doubled up zip codes into separate lines
In [209]:
population[['zip','zip2']]=population.loc[:,'Zip Code'].str.split(
pat =' and ',
expand=True)
population.Population = population.Population.astype(np.float)
population.loc[population.zip2!=None,'Population']=population.loc[population.zip2!=None,'Population']/2
temp_pop = population.loc[population.zip!=None,['Population','zip2']].copy()
temp_pop.rename(columns={'zip2':'zip'},inplace=True)
population = pd.concat([population.loc[:,['Population','zip']],
temp_pop],axis=0)
population.head(2)
Out[209]:
| Population | zip | |
|---|---|---|
| 1 | 53180.0 | 90650 |
| 2 | 52381.0 | 90011 |
Re-run Regression¶
- With population
- Without PM2.5
In [210]:
asthma_aqi = asthma_aqi.merge(population,how='left',on='zip')
y =asthma_aqi.loc[:,'Incidents']
x =asthma_aqi.loc[:,['OZONE','Population']]
x['c'] = 1
glm_model = sm.GLM(y,x,missing='drop',family=sm.families.Poisson())
ols_model2 = sm.OLS(y,x,missing='drop')
glm_results = glm_model.fit()
results = ols_model2.fit()
print(glm_results.summary())
pickle.dump([glm_results,glm_model],open('glm_model_pop_results.p','wb'))
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: Incidents No. Observations: 416
Model: GLM Df Residuals: 413
Model Family: Poisson Df Model: 2
Link Function: log Scale: 1.0000
Method: IRLS Log-Likelihood: -12489.
Date: Thu, 25 Oct 2018 Deviance: 22222.
Time: 13:38:55 Pearson chi2: 2.48e+04
No. Iterations: 5 Covariance Type: nonrobust
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
OZONE 0.0030 0.000 12.910 0.000 0.003 0.003
Population 5.451e-05 2.87e-07 190.139 0.000 5.39e-05 5.51e-05
c 3.8535 0.014 274.430 0.000 3.826 3.881
==============================================================================
Poisson Regression¶
- Summary above was a Poisson regression instead of ols.
- Use GLM() to run a generalized linear model instead of an ols
- Specify the type of regression with family. We have count data, so:
- sm.GLM(y,x,missing='drop',sm.families.Poisson())
- GLMs don't have all the fancy diagnostic plots, so we'll continue with ols results for now.
Partial Regressions¶
In [211]:
fig = plt.figure(figsize=(12,8))
fig = sm.graphics.plot_partregress_grid(results, fig=fig)
Influence plot for outsized-effect of any observations¶
In [212]:
fig, ax = plt.subplots(figsize=(12,8))
fig = sm.graphics.influence_plot(results, ax=ax, criterion="cooks")
Diagnostic plots¶
In [204]:
fig = plt.figure(figsize=(12,8))
fig = sm.graphics.plot_regress_exog(results, "OZONE", fig=fig)
SciKitLearn¶
- Package for machine learning models
- Structured like statsmodels:
- Create a model
- Train on data
- Output results object
- Very good documentation:
Clustering¶
- Learn more about clustering here:
http://scikit-learn.org/stable/modules/clustering.html - Many algorithms, each good in different contexts

K-Means
Randomly pick k initial centroids
K-Means
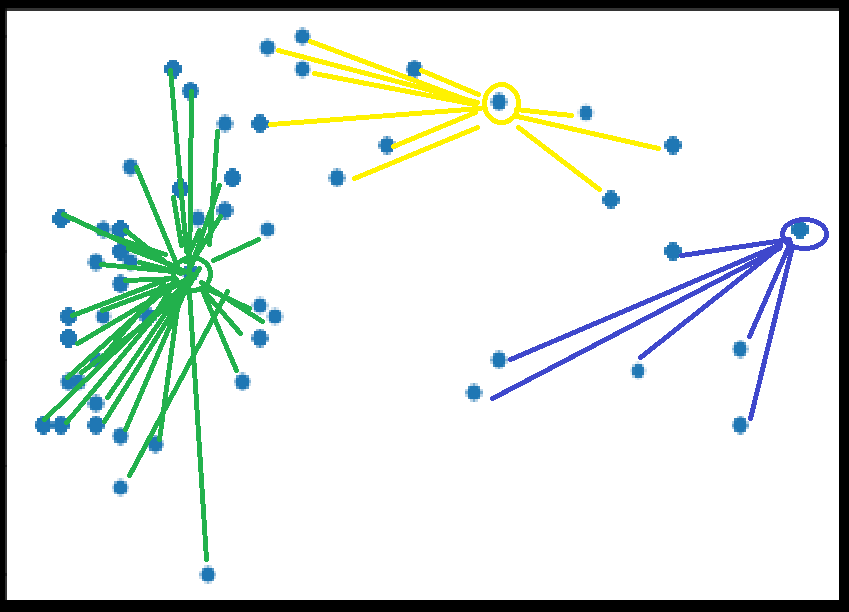
Assign all points to nearest centroids
K-Means
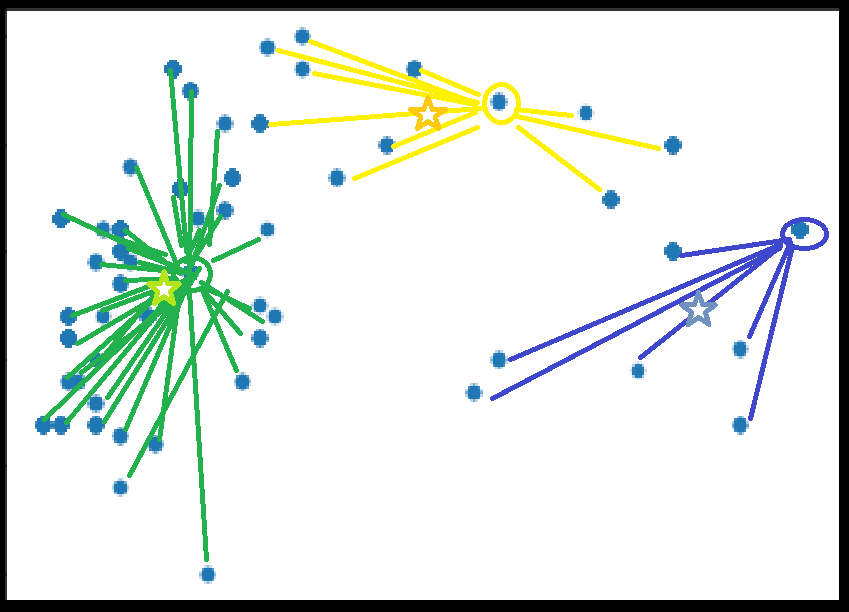
Calculate new centroids for each set
K-Means
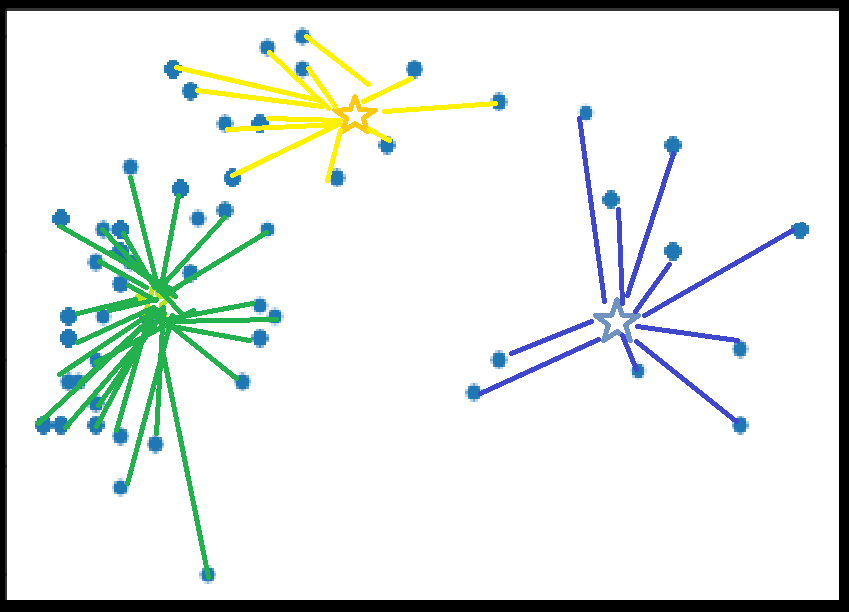
Assign all points to nearest centroid
K-Means

Calculate new centroids, assign points, continue until no change
Prepare data¶
- Statsmodels default drops null values
- Drop rows with missing values first
- Standardize the data so they're all on the same scale
In [325]:
model_df = asthma_aqi.loc[:,['OZONE','PM2.5','Incidents',]]
model_df.dropna(axis=0,inplace=True)
model_df = (model_df - model_df.mean()) / (model_df.max() - model_df.min())
Create and train the model¶
- Initialize a model with three clusters
- fit the model
- extract the labels
In [ ]:
asthma_air_clusters=cluster.KMeans(n_clusters = 3)
asthma_air_clusters.fit(model_df)
model_df['clusters3']=asthma_air_clusters.labels_
Look At Clusters¶
- Our data are very closely clustered, OLS was probably not appropriate.
In [326]:
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure(figsize=(4, 3))
ax = Axes3D(fig, rect=[0, 0, .95, 1], elev=48, azim=134)
labels = asthma_air_clusters.labels_
ax.scatter(model_df.loc[:, 'PM2.5'], model_df.loc[:, 'OZONE'], model_df.loc[:, 'Incidents'],
c=labels.astype(np.float), edgecolor='k')
ax.set_xlabel('Particulates')
ax.set_ylabel('Ozone')
ax.set_zlabel('Incidents')
Out[326]:
Text(0.5,0,'Incidents')
Your Turn¶
- Try a different number of clusters
- Add population data
- Try a different clustering algorithm
Until Next Time¶