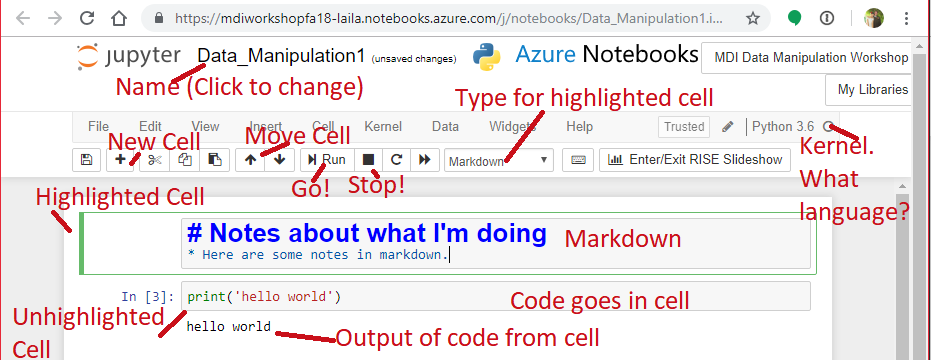
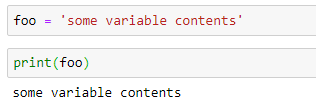
Collecting and Using Data in Python¶
Laila A. Wahedi, PhD¶
Massive Data Institute Postdoctoral Fellow
McCourt School of Public Policy
Follow along:¶
- Slides: http://Wahedi.us, Tutorial
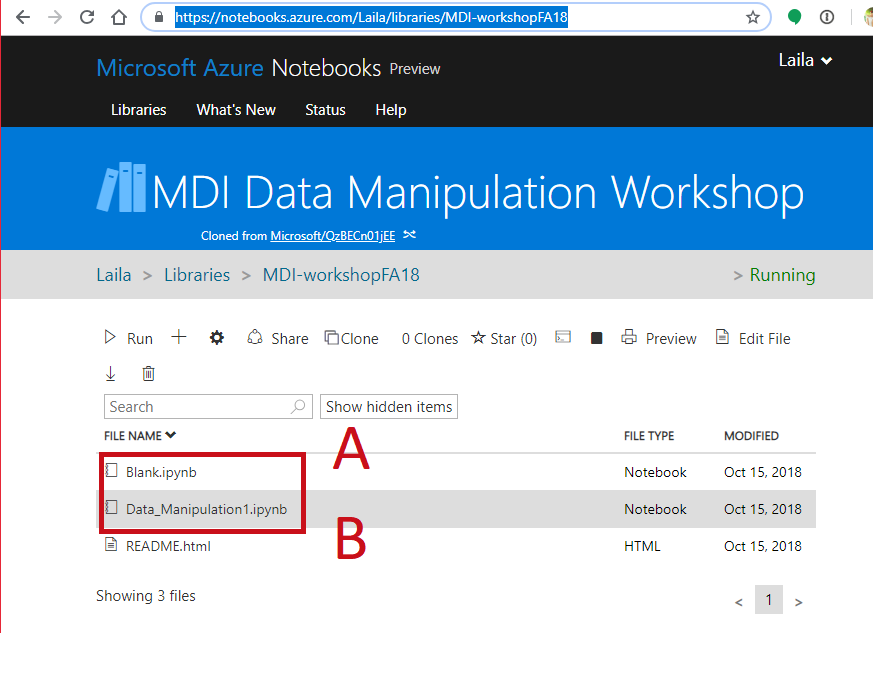
- Interactive Notebook: https://notebooks.azure.com/Laila/libraries/MDI-workshopFA18
Follow Along¶
- Go to https://notebooks.azure.com/Laila/libraries/MDI-workshopFA18
- Clone the directory
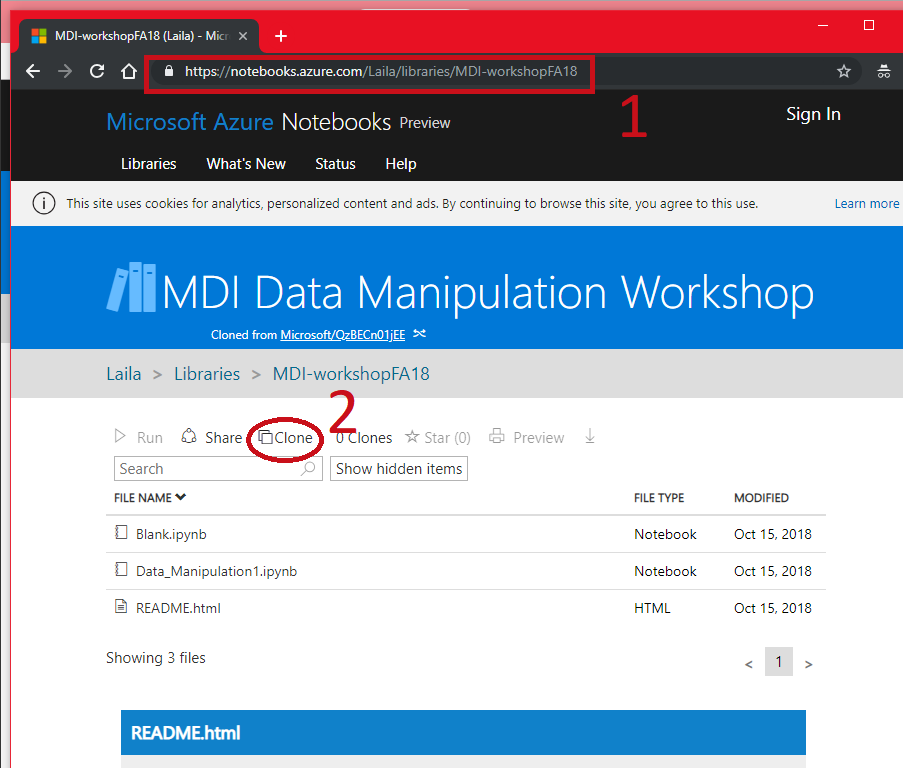
Follow Along¶
- Sign in with any Microsoft Account (Hotmail, Outlook, Azure, etc.)
- Create a folder to put it in, mark as private or public
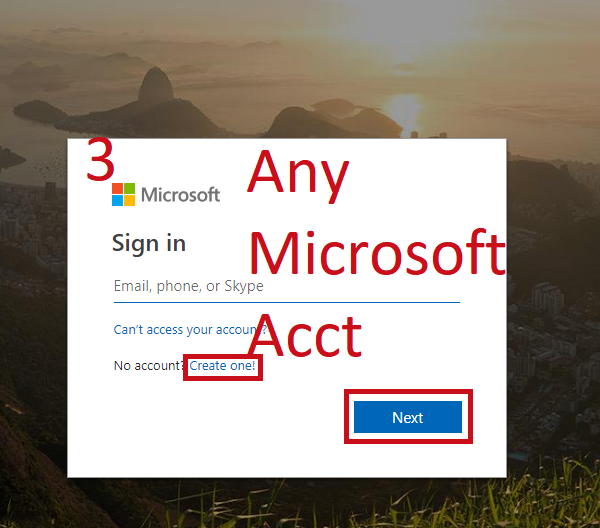
Follow Along¶
- Open a notebook
- Open this notebook to have the code to play with
- Open a blank notebook to follow along and try on your own.
Your Environment¶
- Jupyter Notebook Hosted in Azure
- Want to install it at home?
- Install the Anaconda distribution of Python https://www.anaconda.com/download/
- Install Jupyter Notebooks http://jupyter.org/install
Your Environment: Saving¶
- If your kernel dies, data are gone.
- Not R or Stata, you can't save your whole environment
- Data in memory more than spreadsheets
- Think carefully about what you want to save and how.
Easy Saving (more later)¶
- dump to save the data to hard drive (out of memory)
Contents of the command:
- variable to save,
- File to dump the variable into:
- open(
"name of file in quotes",
"wb") "Write Binary"
- open(
Note: double and single quotes both work
In [4]:
import pickle
mydata = [1,2,3,4,5,6,7,8,9,10]
pickle.dump(mydata, open('mydata.p','wb'))
Save more than one variable:¶
- Put them in a list
In [6]:
more_data = [10,9,8,7,6,5,4,3,2,1]
pickle.dump([mydata,more_data], open('so_much_data.p','wb'))
Loading the data from a pickle¶
- open(<"path to file">
"rb") "Read Binary" - Don't mix up rb and wb. wb will overwrite rb.
In [7]:
mydata = pickle.load(open("mydata.p",'rb'))
print(mydata)
[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
Unpack the variables you saved on the fly¶
In [12]:
[mydata, more_data] = pickle.load(open('so_much_data.p','rb'))
print(mydata)
print(more_data)
[1, 2, 3, 4, 5, 6, 7, 8, 9, 10] [10, 9, 8, 7, 6, 5, 4, 3, 2, 1]
Representing Data¶
- 1D Vectors of data in:
- Lists
- Arrays
- Series
Lists¶
- Square brackets []
- Can contain anything
- Ordered zero-indexing
- Slice with :
- Negatives to go backwards
- Third position to skip
In [119]:
my_list = [1,3,2,4,7,'Sandwich']
print(len(my_list))
print(my_list[0:2])
print(my_list[-1])
print(my_list[0:4:2])
6 [1, 3] Sandwich [1, 2]
Arrays in Numpy¶
- Like arrays from Matlab
- Vectors and multi-dimensional arrays
- Numpy and scipy do math functions, and output in arrays
- Index like lists
In [14]:
import numpy as np
my_array = np.random.poisson(lam=3,size=10)
print(my_array)
print(my_array.shape)
[0 5 4 2 6 2 3 1 2 2] (10,)
Series in Pandas¶
- Pandas is a package that creates labeled data frames
- Series are 1d Vectors
- Instantiate from list or array
- Built on Numpy
In [16]:
import pandas as pd
my_series = pd.Series(my_list)
my_series.shape
Out[16]:
(6,)
Why Series: Label your data¶
In [17]:
my_series = pd.Series(my_array,
index = [1,2,3,'cat','dog','10','n',8,7,6])
print(my_series)
(10,) 1 0 2 5 3 4 cat 2 dog 6 10 2 n 3 8 1 7 2 6 2 dtype: int64
Why Series: Suite of tools¶
In [18]:
print(my_series.mean())
my_series = pd.Series(['hello world','hello planet'])
print(my_series.str.replace('hello','goodbye'))
2.7 0 goodbye world 1 goodbye planet dtype: object
Arrays Series and Lists Can Be Converted¶
In [20]:
new_list = list(my_array)
print(new_list)
[0, 5, 4, 2, 6, 2, 3, 1, 2, 2]
Two Dimensions¶
- List of lists
- Dictionary of lists
- Array
- Pandas Data Frame
Lists of Lists (or tuples)¶
- Tuples are ordered collections like lists, but can't be changed once instantiated.
- Each item in list contains a row.
- Remember the position/order of your variables.
In [24]:
my_2d_list = [[1,4],[2,1],[8,10],[4,7],[9,2],[4,5]]
my_3var_list = [(1,4,7),(2,1,0),(8,10,2),(4,7,4),(9,2,7),(4,5,3)]
Add a variable from another list¶
- You can only add to a list of lists, not tuples
- Must be the proper order and same length
In [25]:
for i,new_var in enumerate(my_list):
my_2d_list[i].append(new_var)
print(my_2d_list)
[[1, 4, 1], [2, 1, 3], [8, 10, 2], [4, 7, 4], [9, 2, 7], [4, 5, 8]]
Keep Track of Variable Names With Dictionaries¶
- Curly Brackets
- Lots of memory, but search columns fast
- Easily add variables
- Index data with labels
In [90]:
my_dict = {
'var1':[1,2,8,4,9,4],
'var2': [4,1,10,7,2,5]
}
my_dict['var3']=my_list
print(my_dict['var3'])
[1, 3, 2, 4, 7, 8]
Use numpy to maintain a matrix shape¶
- Instantiate a 2d array with a list of lists or tuples
- Each variable is a column, each internal list/tuple a row
- Index each dimension like a list, separated by a comma. [row,column]
In [120]:
my_matrix = np.array(my_2d_list)
my_other_matrix = np.array(my_3var_list)
print(my_matrix)
print(my_matrix[0,0:2])
[[ 1 4 1] [ 2 1 3] [ 8 10 2] [ 4 7 4] [ 9 2 7] [ 4 5 8]] [1 4]
Concatenate your matrices by stacking¶
- Axis = 0
In [37]:
big_matrix = np.concatenate([my_matrix, my_other_matrix],axis=0)
print(big_matrix)
[[ 1 4 1] [ 2 1 3] [ 8 10 2] [ 4 7 4] [ 9 2 7] [ 4 5 8] [ 1 4 7] [ 2 1 0] [ 8 10 2] [ 4 7 4] [ 9 2 7] [ 4 5 3]]
Concatenate your matrices side by side¶
- Axis = 1
In [41]:
big_matrix = np.concatenate([my_matrix, my_other_matrix],axis=1)
print(big_matrix)
[[ 1 4 1 1 4 7] [ 2 1 3 2 1 0] [ 8 10 2 8 10 2] [ 4 7 4 4 7 4] [ 9 2 7 9 2 7] [ 4 5 8 4 5 3]]
Do Matrix Operations¶
- Scalar multiplication
- Point-wise addition, subtraction, etc.
- Transpose
In [43]:
print(my_matrix.T + my_other_matrix.T*5)
[[ 6 12 48 24 54 24] [24 6 60 42 12 30] [36 3 12 24 42 23]]
Instantiate A Random Matrix For Simulations¶
- List of distributions here: https://docs.scipy.org/doc/numpy-1.14.0/reference/routines.random.html
In [126]:
my_rand_matrix = np.random.randn(5,3)
print(my_rand_matrix)
[[-1.55580936 -0.17261167 0.75196263] [ 0.84330579 1.33124031 0.3352658 ] [ 0.27599767 -1.15216039 -0.78153232] [-0.05247263 0.60094014 0.60057787] [-1.16768905 -0.43397707 -0.75927943]]
Index like a list with a comma between dimensions:¶
- [row,column]
- Each Column From A Different Normal Distribution:
- Multiply normal distribution by sigma, add mu
In [127]:
my_rand_matrix[:,0]=my_rand_matrix[:,0]*.5+5
my_rand_matrix[:,1]=my_rand_matrix[:,1]*.5-5
my_rand_matrix[:,2]=my_rand_matrix[:,2]*10+50
print(my_rand_matrix.T)
[[ 4.22209532 5.42165289 5.13799884 4.97376368 4.41615548] [ -5.08630584 -4.33437984 -5.5760802 -4.69952993 -5.21698854] [ 57.51962633 53.35265798 42.1846768 56.00577874 42.40720567]]
Sparse Matrices Save Memory When You Have Lots of Zeros¶
- Create a big empty array
- Create indexes to add values
- Add some values to each coordinate. e.g. place 4 in position (1,3,8)
In [128]:
BIG_array = np.zeros((100,100))
rows = (1,6,29,40,43,50)
columns = (3,6,90,58,34,88)
BIG_array[(rows,columns)]=[4,6,14,1,3,22]
Sparse Matrices Save Memory When You Have Lots of Zeros¶
- Turn the matrix into a sparse matrix
- Use scipy package
- Will turn itself back if too big
- Different types good for different things. See: https://docs.scipy.org/doc/scipy/reference/sparse.html
In [129]:
import scipy as sp
from scipy import sparse
BIG_array = sparse.csc_matrix(BIG_array)
print(BIG_array)
(1, 3) 4.0 (6, 6) 6.0 (43, 34) 3.0 (40, 58) 1.0 (50, 88) 22.0 (29, 90) 14.0
Maintain Shape AND Labels with Pandas¶
- DataFrames like R
- Lots of built in functions
- Instantiate from a dictionary...
In [130]:
df = pd.DataFrame(my_dict)
df
Out[130]:
| var1 | var2 | var3 | |
|---|---|---|---|
| 0 | 1 | 4 | 1 |
| 1 | 2 | 1 | 3 |
| 2 | 8 | 10 | 2 |
| 3 | 4 | 7 | 4 |
| 4 | 9 | 2 | 7 |
| 5 | 4 | 5 | 8 |
Instantiate Your Data Frame...¶
- From a list of lists/tuples
In [131]:
df = pd.DataFrame(my_2d_list,
columns = ['var1','var2','var3'])
df
Out[131]:
| var1 | var2 | var3 | |
|---|---|---|---|
| 0 | 1 | 4 | 1 |
| 1 | 2 | 1 | 3 |
| 2 | 8 | 10 | 2 |
| 3 | 4 | 7 | 4 |
| 4 | 9 | 2 | 7 |
| 5 | 4 | 5 | 8 |
Instantiate Your Data Frame...¶
- From a matrix
- Name your rows too!
In [132]:
df = pd.DataFrame(my_rand_matrix,
columns = ['dist_1','dist_2','dist_3'],
index = ['obs1','obs2','obs3','obs4','fred'])
df
Out[132]:
| dist_1 | dist_2 | dist_3 | |
|---|---|---|---|
| obs1 | 4.222095 | -5.086306 | 57.519626 |
| obs2 | 5.421653 | -4.334380 | 53.352658 |
| obs3 | 5.137999 | -5.576080 | 42.184677 |
| obs4 | 4.973764 | -4.699530 | 56.005779 |
| fred | 4.416155 | -5.216989 | 42.407206 |
Summarize Your Data¶
In [133]:
df.describe()
Out[133]:
| dist_1 | dist_2 | dist_3 | |
|---|---|---|---|
| count | 5.000000 | 5.000000 | 5.000000 |
| mean | 4.834333 | -4.982657 | 50.293989 |
| std | 0.501574 | 0.479122 | 7.452384 |
| min | 4.222095 | -5.576080 | 42.184677 |
| 25% | 4.416155 | -5.216989 | 42.407206 |
| 50% | 4.973764 | -5.086306 | 53.352658 |
| 75% | 5.137999 | -4.699530 | 56.005779 |
| max | 5.421653 | -4.334380 | 57.519626 |
Look at your data with Matplotlib integration¶
- Matplotlib is like plotting in matlab
- Try ggplot package for ggplot2 in python
- See also Seaborn and Plotly
- Use some ipython magic to see plots inline
In [134]:
import matplotlib.pyplot as plt
%matplotlib inline
df.plot.density()
Out[134]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdaf0662128>
One Variable At A Time:¶
In [135]:
df.dist_1.plot.hist(bins=3)
Out[135]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdaf06b4828>
Real Data¶
Load Data from a text file¶
- Start by googling it: http://lmgtfy.com/?q=pandas+load+csv
- Same method for comma (csv), tab (tab), |, and other separators
- Excel and R can both output spreadsheets to csv
We will use the Big Allied and Dangerous Data from START¶
In [95]:
baad_covars = pd.read_csv('BAAD_1_Lethality_Data.tab',sep='\t')
Look at the data¶
- Also try .tail()
In [96]:
baad_covars.head(3)
Out[96]:
| mastertccode3606 | group | statespond | cowmastercountry | masterccode | fatalities19982005 | OrgAge | ordsize | terrStrong | degree | ContainRelig | ContainEthno | LeftNoReligEthno | PureRelig | PureEthno | ReligEthno | ContainRelig2 | ContainEthno2 | Islam | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 50 | Animal Liberation Front (ALF) | 0 | United States of America | 2 | 0 | 30 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 89 | Army of God | 0 | United States of America | 2 | 1 | 24 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| 2 | 113 | Cambodian Freedom Fighters (CFF) | 0 | United States of America | 2 | 0 | 8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
Explore the data structure¶
In [76]:
print(baad_covars.shape)
baad_covars.columns
(395, 19)
Out[76]:
Index(['mastertccode3606', 'group', 'statespond', 'cowmastercountry',
'masterccode', 'fatalities19982005', 'OrgAge', 'ordsize', 'terrStrong',
'degree', 'ContainRelig', 'ContainEthno', 'LeftNoReligEthno',
'PureRelig', 'PureEthno', 'ReligEthno', 'ContainRelig2',
'ContainEthno2', 'Islam'],
dtype='object')
Rename things and adjust values¶
- Use dictionaries to rename and replace
In [77]:
baad_covars.rename(columns = {'cowmastercountry':'country',
'masterccode':'ccode',
'mastertccode3606':'group_code',
'fatalities19982005':'fatalities'},
inplace = True)
baad_covars.replace({'country':{'United States of America':'US'}},
inplace = True)
print('Dimensions: ',baad_covars.shape)
baad_covars.head()
Dimensions: (395, 19)
Out[77]:
| group_code | group | statespond | country | ccode | fatalities | OrgAge | ordsize | terrStrong | degree | ContainRelig | ContainEthno | LeftNoReligEthno | PureRelig | PureEthno | ReligEthno | ContainRelig2 | ContainEthno2 | Islam | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 50 | Animal Liberation Front (ALF) | 0 | US | 2 | 0 | 30 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 89 | Army of God | 0 | US | 2 | 1 | 24 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| 2 | 113 | Cambodian Freedom Fighters (CFF) | 0 | US | 2 | 0 | 8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 3 | 126 | Coalition to Save the Preserves (CSP) | 0 | US | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 153 | Earth Liberation Front (ELF) | 0 | US | 2 | 0 | 14 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Set a useful index¶
In [39]:
#Set the index
baad_covars.set_index(['group_code'],inplace = True)
baad_covars.head()
Out[39]:
| group | statespond | country | ccode | fatalities | OrgAge | ordsize | terrStrong | degree | ContainRelig | ContainEthno | LeftNoReligEthno | PureRelig | PureEthno | ReligEthno | ContainRelig2 | ContainEthno2 | Islam | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| group_code | ||||||||||||||||||
| 50 | Animal Liberation Front (ALF) | 0 | US | 2 | 0 | 30 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 89 | Army of God | 0 | US | 2 | 1 | 24 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| 113 | Cambodian Freedom Fighters (CFF) | 0 | US | 2 | 0 | 8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 126 | Coalition to Save the Preserves (CSP) | 0 | US | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 153 | Earth Liberation Front (ELF) | 0 | US | 2 | 0 | 14 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Save Your Changes¶
- Save it to a usable spreadsheet instead of an unreadable binary
In [78]:
baad_covars.to_csv('updated_baad.csv')
Slicing¶
- Get specific values from the dataframe.
- Pandas has several slice operators.
- iloc can be used to index the row by ordered integer. i.e. first row is 0, second row is 1, etc. Use this option sparingly. Better practice to use the index you have created.
- loc uses the named index and columns.
- Index using [row, columns]
- Put your column names in a list
- Use : for all values
- Notice that the output keeps the index names.
In [40]:
baad_covars.loc[:, ['fatalities']].head()
Out[40]:
group_code 50 0 89 1 113 0 126 0 153 0 Name: fatalities, dtype: int64
Slicing Using Conditionals¶
- Put conditionals in parentheses
- Stack multiple conditionals using:
- & when both conditions must always apply
- | when at least one condition must apply
In [41]:
baad_covars.loc[(baad_covars.fatalities>1) | (baad_covars.degree>=1),
['group','country']].head()
Out[41]:
| group | country | |
|---|---|---|
| group_code | ||
| 50 | Animal Liberation Front (ALF) | US |
| 153 | Earth Liberation Front (ELF) | US |
| 30035 | Maras Salvatruchas | US |
| 10042 | Group of Guerilla Combatants of Jose Maria Mor... | Mexico |
| 246 | Justice Army of the Defenseless People | Mexico |
Find a list of religious groups with territory¶
Find a list of religious groups with territory¶
In [102]:
baad_covars.loc[(baad_covars.ContainRelig==1)&
(baad_covars.terrStrong==1),['group']]
Out[102]:
| group | |
|---|---|
| 198 | Hisba |
| 203 | Lord's Resistance Army (LRA) |
| 238 | Ansar al-Islam |
| 252 | Mahdi Army |
| 272 | Hezbollah |
| 281 | Hamas |
| 298 | Hizb-I-Islami |
| 303 | Taliban |
| 305 | Islamic Movement of Uzbekistan (IMU) |
| 356 | al-Qaeda |
| 379 | Abu Sayyaf Group (ASG) |
| 383 | Moro Islamic Liberation Front (MILF) |
| 384 | Moro National Liberation Front (MNLF) |
| 391 | Jemaah Islamiya (JI) |
| 392 | Laskar Jihad |
Plot a histogram of organization age with 20 bins¶
Plot a histogram of organization age with 20 bins¶
In [103]:
baad_covars.OrgAge.plot.hist(bins=10)
Out[103]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdaf0883588>
Grouping By Variables¶
- Groupby(): List the variables to group by
- .function(): How to aggregate the rows
- Try: .count(), .mean(), .first(), .mode()
In [83]:
state_level = baad_covars.loc[:,['country','OrgAge',
'ordsize','degree',
'fatalities']
].groupby(['country']).sum()
state_level.head()
Out[83]:
| OrgAge | ordsize | degree | fatalities | |
|---|---|---|---|---|
| country | ||||
| Afghanistan | 58 | 4 | 11 | 353 |
| Algeria | 24 | 2 | 6 | 409 |
| Angola | 83 | 5 | 0 | 276 |
| Argentina | 2 | 0 | 0 | 0 |
| Bangladesh | 83 | 5 | 4 | 80 |
In [62]:
baad_covars['big'] = 0
baad_covars.loc[(baad_covars.fatalities>1) |
(baad_covars.degree>=1),
'big']=1
baad_covars.big.head()
Out[62]:
group_code 50 1 89 0 113 0 126 0 153 1 Name: big, dtype: int64
In [54]:
print(type(np.nan))
baad_covars.loc[(baad_covars.fatalities>1) | (baad_covars.degree>=1),
['terrStrong']] = None
baad_covars.loc[(baad_covars.fatalities>1) | (baad_covars.degree>=1),
['terrStrong']].head()
<class 'float'>
Out[54]:
| terrStrong | |
|---|---|
| group_code | |
| 50 | NaN |
| 153 | NaN |
| 30035 | NaN |
| 10042 | NaN |
| 246 | NaN |
In [56]:
baad_covars.loc[baad_covars.terrStrong.isnull(),'terrStrong'].head()
Out[56]:
group_code 50 NaN 153 NaN 30035 NaN 10042 NaN 246 NaN Name: terrStrong, dtype: float64
In [58]:
baad_covars['terrStrong'] = baad_covars.terrStrong.fillna(-77)
baad_covars.terrStrong.head()
Out[58]:
group_code 50 -77.0 89 0.0 113 0.0 126 0.0 153 -77.0 Name: terrStrong, dtype: float64
In [60]:
baad_covars_dropped = baad_covars.dropna(axis='index',
subset=['terrStrong'],
inplace=False)
Reindexing: Pop the index out without losing it¶
In [63]:
baad_covars.reset_index(inplace=True,
drop = False)
baad_covars.head()
Out[63]:
| group_code | group | statespond | country | ccode | fatalities | OrgAge | ordsize | terrStrong | degree | ContainRelig | ContainEthno | LeftNoReligEthno | PureRelig | PureEthno | ReligEthno | ContainRelig2 | ContainEthno2 | Islam | big | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 50 | Animal Liberation Front (ALF) | 0 | US | 2 | 0 | 30 | 0 | -77.0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1 | 89 | Army of God | 0 | US | 2 | 1 | 24 | 0 | 0.0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2 | 113 | Cambodian Freedom Fighters (CFF) | 0 | US | 2 | 0 | 8 | 0 | 0.0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3 | 126 | Coalition to Save the Preserves (CSP) | 0 | US | 2 | 0 | 6 | 0 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 153 | Earth Liberation Front (ELF) | 0 | US | 2 | 0 | 14 | 0 | -77.0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Set a multi-index¶
- Order Matters. What happens when you reverse group and country?
In [64]:
baad_covars.set_index(['group','country'],inplace = True)
baad_covars.head()
Out[64]:
| group_code | statespond | ccode | fatalities | OrgAge | ordsize | terrStrong | degree | ContainRelig | ContainEthno | LeftNoReligEthno | PureRelig | PureEthno | ReligEthno | ContainRelig2 | ContainEthno2 | Islam | big | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| group | country | ||||||||||||||||||
| Animal Liberation Front (ALF) | US | 50 | 0 | 2 | 0 | 30 | 0 | -77.0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Army of God | US | 89 | 0 | 2 | 1 | 24 | 0 | 0.0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Cambodian Freedom Fighters (CFF) | US | 113 | 0 | 2 | 0 | 8 | 0 | 0.0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Coalition to Save the Preserves (CSP) | US | 126 | 0 | 2 | 0 | 6 | 0 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Earth Liberation Front (ELF) | US | 153 | 0 | 2 | 0 | 14 | 0 | -77.0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Did you get an error?¶
- Don't forget to reset the index first!
- Go ahead and change it back for the next step.
Using the new index, make a new dataframe¶
- Note the new slicing operator for multi-index
In [65]:
indonesia_grps = baad_covars.xs('Indonesia',level = 'country',drop_level=False)
indonesia_grps = indonesia_grps.loc[indonesia_grps.fatalities>=1,['degree','ContainRelig',
'ContainEthno','terrStrong',
'ordsize','OrgAge']]
indonesia_grps.head()
Out[65]:
| degree | ContainRelig | ContainEthno | terrStrong | ordsize | OrgAge | ||
|---|---|---|---|---|---|---|---|
| group | country | ||||||
| Free Aceh Movement (GAM) | Indonesia | 1 | 1 | 1 | -77.0 | 2 | 31 |
| Jemaah Islamiya (JI) | Indonesia | 2 | 1 | 0 | -77.0 | 1 | 13 |
| Laskar Jihad | Indonesia | 3 | 1 | 1 | -77.0 | 0 | 6 |
| South Maluku Republic (RMS) | Indonesia | 0 | 0 | 1 | -77.0 | 2 | 8 |
Warning: Making copies¶
- If you set a variable as equal to an object, Python creates a reference rather than copying the whole object. More efficient, unless you really want to make a copy
In [137]:
little_df = pd.DataFrame([1,2,3,4,5],columns = ['A'])
little_df['B']=[0,1,0,1,1]
copied_df = little_df
print('before:')
print(copied_df)
little_df.loc[little_df.A == 3,'B'] = 'Sandwich'
print('after')
print(copied_df)
before: A B 0 1 0 1 2 1 2 3 0 3 4 1 4 5 1 after A B 0 1 0 1 2 1 2 3 Sandwich 3 4 1 4 5 1
What happened?¶
- copied_df changed when little_df changed.
- Let's fix that: import "copy"
In [139]:
import copy
little_df = pd.DataFrame([1,2,3,4,5],columns = ['A'])
little_df['B']=[0,1,0,1,1]
copied_df = little_df.copy()
print('before:')
print(copied_df)
little_df.loc[little_df.A == 3,'B'] = 'Sandwich'
print('after')
print(copied_df)
before: A B 0 1 0 1 2 1 2 3 0 3 4 1 4 5 1 after A B 0 1 0 1 2 1 2 3 0 3 4 1 4 5 1
Merging and Concatenating¶
- Merges automatically if shared index
In [140]:
C = pd.DataFrame(['apple','orange','grape','pear','banana'],
columns = ['C'],
index = [2,4,3,0,1])
little_df['C'] = C
little_df
Out[140]:
| A | B | C | |
|---|---|---|---|
| 0 | 1 | 0 | pear |
| 1 | 2 | 1 | banana |
| 2 | 3 | Sandwich | apple |
| 3 | 4 | 1 | grape |
| 4 | 5 | 1 | orange |
Joins¶
- Same as SQL, inner and outer
In [141]:
C = pd.DataFrame(['apple','orange','grape','apple'],
columns = ['C'],
index = [2,4,3,'a'])
C['cuts']=['slices','wedges','whole','spirals']
print('C:')
print(C)
print('Inner: Intersection')
print(little_df.merge(right=C,
how='inner',
on=None,
left_index = True,
right_index =True))
print('Outer: Keep all rows')
print(little_df.merge(right=C,
how='outer',
on=None,
left_index = True,
right_index =True))
print('Left: Keep little_df')
print(little_df.merge(right=C,
how='left',
on=None,
left_index = True,
right_index =True))
print('Right: Keep C')
print(little_df.merge(right=C,
how='right',
on=None,
left_index = True,
right_index =True))
print('Outer, merging on column instead of index')
print(little_df.merge(right=C,
how='outer',
on='C',
left_index = False,
right_index =False))
C:
C cuts
2 apple slices
4 orange wedges
3 grape whole
a apple spirals
Inner: Intersection
A B C_x C_y cuts
2 3 Sandwich apple apple slices
3 4 1 grape grape whole
4 5 1 orange orange wedges
Outer: Keep all rows
A B C_x C_y cuts
0 1.0 0 pear NaN NaN
1 2.0 1 banana NaN NaN
2 3.0 Sandwich apple apple slices
3 4.0 1 grape grape whole
4 5.0 1 orange orange wedges
a NaN NaN NaN apple spirals
Left: Keep little_df
A B C_x C_y cuts
0 1 0 pear NaN NaN
1 2 1 banana NaN NaN
2 3 Sandwich apple apple slices
3 4 1 grape grape whole
4 5 1 orange orange wedges
Right: Keep C
A B C_x C_y cuts
2 3.0 Sandwich apple apple slices
4 5.0 1 orange orange wedges
3 4.0 1 grape grape whole
a NaN NaN NaN apple spirals
Outer, merging on column instead of index
A B C cuts
0 1 0 pear NaN
1 2 1 banana NaN
2 3 Sandwich apple slices
3 3 Sandwich apple spirals
4 4 1 grape whole
5 5 1 orange wedges
/opt/conda/lib/python3.6/site-packages/pandas/core/indexes/base.py:3772: RuntimeWarning: '<' not supported between instances of 'int' and 'str', sort order is undefined for incomparable objects return this.join(other, how=how, return_indexers=return_indexers)
Concatenate¶
- Stack dataframes on top of one another
- Stack dataframes beside one another
In [142]:
add_df = pd.DataFrame({'A':[6],'B':[7],'C':'peach'},index= ['p'])
little_df = pd.concat([little_df,add_df])
little_df
Out[142]:
| A | B | C | |
|---|---|---|---|
| 0 | 1 | 0 | pear |
| 1 | 2 | 1 | banana |
| 2 | 3 | Sandwich | apple |
| 3 | 4 | 1 | grape |
| 4 | 5 | 1 | orange |
| p | 6 | 7 | peach |
Some New Messy Data: Asthma by Zip Code¶
- From California Health and Human Services
https://data.chhs.ca.gov/dataset/asthma-emergency-department-visit-rates-by-zip-code - Note: old version of data
In [107]:
asthma_data = pd.read_csv('asthma-emergency-department-visit-rates-by-zip-code.csv')
asthma_data.head(2)
Out[107]:
| Year | ZIP code | Age Group | Number of Visits | Age-adjusted rate | County Fips code | County | |
|---|---|---|---|---|---|---|---|
| 0 | 2015 | 90004\n(34.07646, -118.309453) | Children (0-17) | 117.0 | 91.7 | 6037 | LOS ANGELES |
| 1 | 2015 | 90011\n(34.007055, -118.258872) | Children (0-17) | 381.0 | 102.8 | 6037 | LOS ANGELES |
Look at those zip codes!¶
Clean Zip Code¶
- We don't need the latitude and longitude
- Create two variables by splitting the zip code variable:
- index the data frame to the zip code variable
- split it in two: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.str.split.html
- assign it to another two variables
- Remember: can't run this cell twice without starting over
In [108]:
asthma_data[['zip','coordinates']] = asthma_data.loc[:,'ZIP code'].str.split(
pat='\n',expand=True)
asthma_data.drop('ZIP code', axis=1,inplace=True)
asthma_data.head(2)
Out[108]:
| Year | Age Group | Number of Visits | Age-adjusted rate | County Fips code | County | zip | coordinates | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2015 | Children (0-17) | 117.0 | 91.7 | 6037 | LOS ANGELES | 90004 | (34.07646, -118.309453) |
| 1 | 2015 | Children (0-17) | 381.0 | 102.8 | 6037 | LOS ANGELES | 90011 | (34.007055, -118.258872) |
Rearrange The Data: Group By¶
- Make child and adult separate columns rather than rows.
- Must specify how to aggregate the columns
https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.groupby.html
In [109]:
asthma_grouped = asthma_data.groupby(by=['Year','zip']).sum()
asthma_grouped.head(4)
Out[109]:
| Number of Visits | Age-adjusted rate | County Fips code | ||
|---|---|---|---|---|
| Year | zip | |||
| 2009 | 90001 | 818.0 | 226.074245 | 18111 |
| 90002 | 836.0 | 265.349315 | 18111 | |
| 90003 | 1542.0 | 369.202131 | 18111 | |
| 90004 | 580.0 | 145.538276 | 18111 |
Lost Columns! Fips summed!¶
Group by: Cleaning Up¶
- Lost columns you can't sum
- took sum of fips
- Must add these back in
- Works because temp table has same index
In [110]:
asthma_grouped.drop('County Fips code',axis=1,inplace=True)
temp_grp = asthma_data.groupby(by=['Year','zip']).first()
asthma_grouped[['fips',
'county',
'coordinates']]=temp_grp.loc[:,['County Fips code',
'County',
'coordinates']].copy()
asthma_grouped.loc[:,'Number of Visits']=\
asthma_grouped.loc[:,'Number of Visits']/2
asthma_grouped.head(2)
Out[110]:
| Number of Visits | Age-adjusted rate | fips | county | coordinates | ||
|---|---|---|---|---|---|---|
| Year | zip | |||||
| 2009 | 90001 | 409.0 | 226.074245 | 6037 | LOS ANGELES | (33.973252, -118.249154) |
| 90002 | 418.0 | 265.349315 | 6037 | LOS ANGELES | (33.949079, -118.247877) |
Rearrange The Data: Pivot¶
- Use pivot and melt to to move from row identifiers to column identifiers and back
https://pandas.pydata.org/pandas-docs/stable/reshaping.html#reshaping-by-melt - Tell computer what to do with every cell:
- Index: Stays the same
- Columns: The column containing the new column labels
- Values: The column containing values to insert
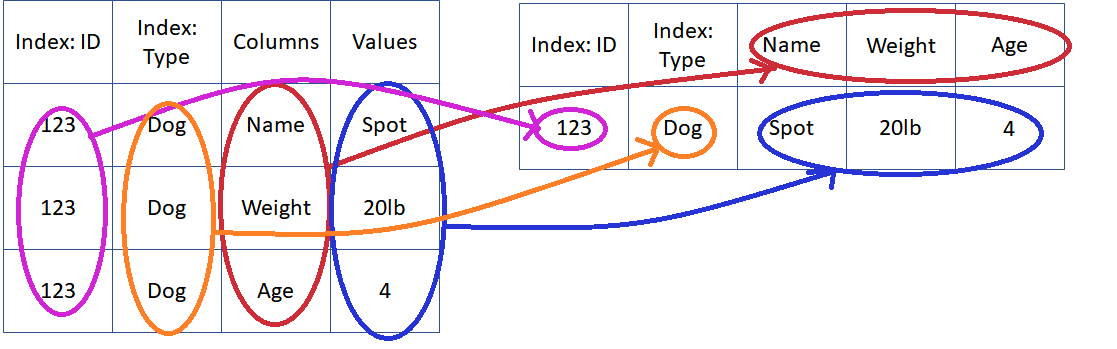
Rearrange The Data: Pivot¶
- Tell computer what to do with every cell:
- Index: Stays the same
- Columns: The column containing the new column labels
- Values: The column containing values to insert
In [111]:
asthma_unstacked = asthma_data.pivot_table(index = ['Year',
'zip',
'County',
'coordinates',
'County Fips code'],
columns = 'Age Group',
values = 'Number of Visits')
asthma_unstacked.reset_index(drop=False,inplace=True)
asthma_unstacked.head(2)
Out[111]:
| Age Group | Year | zip | County | coordinates | County Fips code | Adults (18+) | All Ages | Children (0-17) |
|---|---|---|---|---|---|---|---|---|
| 0 | 2009 | 90001 | LOS ANGELES | (33.973252, -118.249154) | 6037 | 206.0 | 409.0 | 203.0 |
| 1 | 2009 | 90002 | LOS ANGELES | (33.949079, -118.247877) | 6037 | 204.0 | 418.0 | 214.0 |
Rename Columns, Subset Data¶
In [113]:
asthma_unstacked.rename(columns={
'zip':'Zip',
'coordinates':'Coordinates',
'County Fips code':'Fips',
'Adults (18+)':'Adults',
'All Ages':'Incidents',
'Children (0-17)': 'Children'
},
inplace=True)
asthma_2015 = asthma_unstacked.loc[asthma_unstacked.Year==2015,:]
asthma_2015.head(2)
Out[113]:
| Age Group | Year | Zip | County | Coordinates | Fips | Adults | Incidents | Children |
|---|---|---|---|---|---|---|---|---|
| 4693 | 2015 | 90001 | LOS ANGELES | (33.973252, -118.249154) | 6037 | 229.0 | 441.0 | 212.0 |
| 4694 | 2015 | 90002 | LOS ANGELES | (33.949079, -118.247877) | 6037 | 249.0 | 476.0 | 227.0 |
Save Your Data¶
Save Your Data¶
In [114]:
asthma_2015.to_csv('asthma_2015.csv')
See You Next Week!¶
<img src = "https://media.giphy.com/media/6VWz7mToYWdDNYglYr/giphy.gif">